| Detailed Information for Phage Ampersand |
| Discovery Information |
| Found By | Ami Vodi |
| Year Found | 2019 |
| Location Found | Lanham, MD United States |
| Finding Institution | University of Maryland, Baltimore County |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | No |
| GPS Coordinates | 38.989868 N, 76.845759 W Map |
| Discovery Notes | From dry soil, near a deer foot print, collected 5/27/19 |
| Naming Notes | Ampersand is the name of "&" and I thought that it would be interesting and unique as a name |
| Sequencing Information |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Oct 29, 2019 |
| Sequencing Facility | NC State Genomic Sciences Laboratory |
| Shotgun Sequencing Method | Illumina Sequencing |
| Genome length (bp) | 164235 |
| GC Content | 37.8% |
| Character of genome ends | Direct Terminal Repeat |
| End Determination Notes | Examination of reads |
| Fasta file available? | Yes: Download fasta file |
| Characterization |
| Cluster | Unassigned |
| Subcluster | -- |
| Other Cluster Members |
|
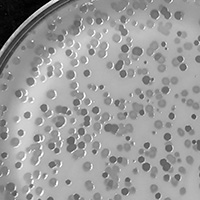
| Plaque Notes | circular, 1-2mm, typically 1.5 mm, after 24 hours at 30 C, turbid plaques, similar to a bull's eye |
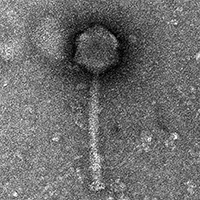
| Morphotype | Myoviridae |
| Final annotation complete? | No |
| Has been Phamerated? | No |
| Uploaded to GenBank? | No |
| Available Files |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |