| Detailed Information for Phage GypsyDanger | |
| Discovery Information | |
| Found By | Ahmed Farshori, William Blankenship |
| Year Found | 2013 |
| Location Found | Baltimore, MD United States |
| Finding Institution | University of Maryland, Baltimore County |
| Program | Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science |
| From enriched soil sample? | Yes |
| GPS Coordinates | 39.25455 N, 76.703453 W Map |
| Discovery Notes | The phage GypsyDanger was discovered in soil sample from a stream running across the periphery of the University of Maryland Baltimore County campus. Soil was extracted from approximately 6cm below the damp stream bank very close to the impervious surface of a parking lot. |
| Naming Notes | The name GypsyDanger was chosen because it is something easy and fun to relate to for the students and professors this year. |
| Sequencing Information | |
| Sequencing Complete? | Yes |
| Date Sequencing Completed | Jan 13, 2014 |
| Sequencing Facility | NC State Genomic Sciences Laboratory |
| Shotgun Sequencing Method | Illumina Sequencing |
| Approximate Shotgun Coverage | 3187 |
| Genome length (bp) | 164029 |
| GC Content | 37.6% |
| Character of genome ends | Direct Terminal Repeat |
| Direct Terminal Repeat Length | 2293 bp |
| End Determination Notes | Buildup of Illumina sequencing reads was clear. |
| Fasta file available? | Yes: Download fasta file |
| Characterization | |
| Cluster | Unassigned |
| Subcluster | -- |
| Other Cluster Members |
Click to ViewA1B2 A1Cooties A5K94 AABSAS Aarhus Aarib Abel235 AbeLyticoln Abora Absfotbb ABTN19 AceMooncake Achilles Actcat127 Adelynn ADES AdEx Adku AdoptMe Advas Aegis0001 Aestusestus1 Ajax AK37 AKisBest Aladdin Alastair Aldie Alfred AliceH2012 Alissa AllMyCellsRDed AlyDip Amicus Ampersand Anaconda Anaisshepherd Anchovy Anders Angel AngeriGhost Anthracilles AOK13 AOKp AP50 Apollo Appalachia ApplePie ApplePieA101 AR1951 Araneum Arash Araxxor ArchibaldCrave Archie14 Argon Aries Arini ARTA Arvgarden ASAPherg Ashina Aspen2314 Asperygus Asphyxia Aster Asterix AstralM Atlee Attesideifagi Aubrey Avani0322 Avatar Aviola Avocado Awesomesauce1 Aycee Azalea Azaleja BA4ever Baanoo Babo BaBooBoo Babyblaze BabyBoo Babybutterfish BabygotBac BacBender BacillaKilla BadBK BadBunnyBaby Bae0001 Baege Baguette BaharaJr BaharaJr2 BAIRD1 Baker BALL3R BALL3R1 Balla Ballerina429 Ballerine Ballislife Balthazaar Balthazar Bastille BC4U Bcp1 Bhoosica BigBertha BM10 ChannelFever ChurroBlast Cletus DingnCase Dingus Doofinshmirtz Elmer Escavirius Freefood FreightTrain Grumio GypsyDanger Helga Herbert HoodyT Hudslayer Hydralpha Ilona JoeB Joonam Kota Lilihipha MightyMouse Momo Natp Oregano Parsons Phabio Rex16 STSebastian Terra Typhen WPh |
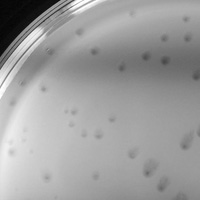
| Plaque Notes | GypsyDanger forms standard lytic plaques with one unique feature; if the plaques are incubated longer than 24 hours the larger ones will form distinct haloes around them that all skew in a single direction, resulting in a "falling meteor" shape. These plaques have been reproduced multiple times and are an anomaly, as other phages in the same lab do not produce such distinct, unidirectionally skewed plaques. |
| Morphotype | Myoviridae |
| Final annotation complete? | No |
| Has been Phamerated? | No |
| Uploaded to GenBank? | No |
| Available Files | |
| Plaque Picture | Download |
| Restriction Digest Picture | Download |
| EM Picture | Download |